Substance use disorders (SUDs), including drug, alcohol and nicotine dependence, are global threats for physical and mental public health. Their estimated prevalence lies in range from ~4% (for drug use disorder) to >20% (for smoking). This group of disorders is particularly relevant for low- and middle-income countries. Thus, in Ukraine, according to WHO and UN data and Ministry of Health statistics, ~350,000 people inject drugs 1, around 6% of adult population abuse alcohol 2, and 23.7% smoke daily 3. Drug abuse frequently leads to HIV, HCV and HBV infection, deaths from overdose, multiple psychiatric, metabolic and cardiovascular events, as well as social problems. Across Eastern Europe and Baltia (Ukraine, Belarus, Russia, Lithuania, Latvia and Estonia), up to one-third of deaths are attributed to alcohol consumption 4. Over 80% of the world's 1.3 billion tobacco users live in low- and middle-income countries 2. Taken together, substance abuse annually leads to millions of deaths worldwide 2.
In spite of dozens of genome-wide association studies (GWASs) of SUDs that have been carried out in the recent years, there are still only a few loci reported at the genome-wide significance outside the ADH gene cluster for alcohol and the CHRNA5-CHRNA3-CHRNB4 gene cluster for nicotine dependence. In addition, to our knowledge, no GWASs of SUDs have been carried out in Eastern Europe, except for a GWAS of alcohol consumption in Russians, that gave suggestive evidence of different genetic architecture of SUDs 5 in Eastern Europe, along with some data from candidate gene studies in Roma populations 6, 7 and our preliminary data 8,9. Another important problem related to SUDs is their high levels of co-morbidity.
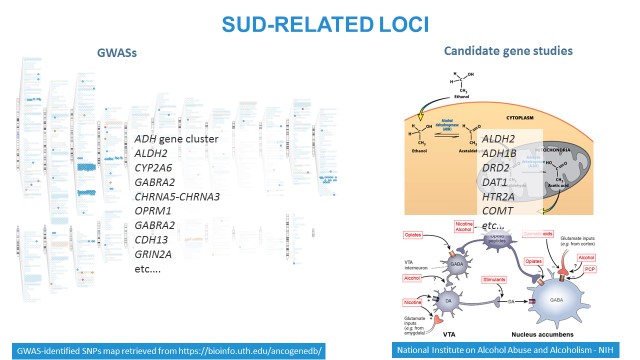
There are two traditional approaches to identify and replicate genomic variations associated with complex phenotypes, such as SUDs: GWASs and candidate gene studies. The left panel: GWAS signals mapped on chromosomes. The upper right panel: variations in the loci involved in substance metabolism (on the example of alcohol) are associated with liability to dependence. The lower right panel: variations affecting neurobiologic pathways, such as reward system, may be also associated with substance abuse or dependence.
We propose a multivariate approach to candidate gene study of SUDs in Ukraine. We collect DNA samples of individuals, living in Ukraine, with the data regarding their SUDs-related diagnoses, substance consumption, and some psychological scales. The study is approved by a local Ethics Committee, all individuals give their informed consents for participation in the study, and the data is anonymised. We aim to genotype at least 500 samples overall, with the desired number of individuals reaching 700 to increases the study power.
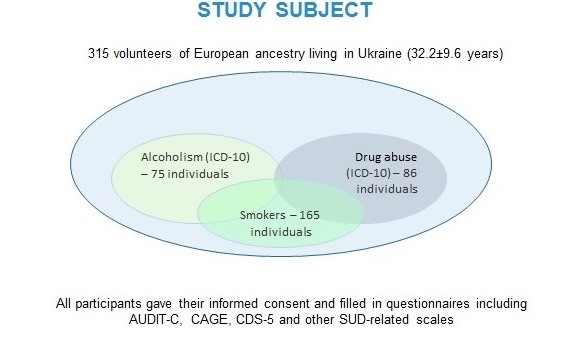
Our preliminary data on 315 individuals suggest some associations of genetic loci implicated in substance metabolism and neurobiological effects with alcohol and nicotine dependence 8, 9, moreover, some of the associations seem to be pleiotropic, i.e. significant regarding to two phenotypes (unpublished; the data were presented at ENCODS-2019). However, we cannot draw any reliable conclusions, since our study is yet underpowered and includes fewer than 100 participants in drug and alcohol dependent groups.
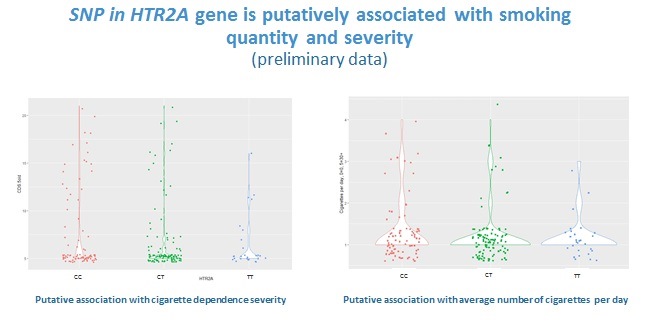
As an outcome of the proposed candidate gene study, we hope to get results that would shed light on effects and possible pleiotropic effects of the tested loci on SUDs, in Ukrainian population. To our knowledge, this is the first genetic association study of this group of phenotypes in Ukraine. Further we plan to apply for funding to cover a multivariate GWAS, which would allow to extract information on genetic correlation with other mental health disorders and on creation of a polygenic risk score relevant for Ukrainian population.
References
1 https://wdr.unodc.org/wdr2019/en/exsum.html
2 https://www.who.int
3 Global Adult Tobacco Survey (GATS). Report Ukraine 2017.
4 Ritchie, Roser. Alcohol consumption, in: Our world in data, 2018.
5 Nizamutdinov et al. bioRxiv 2018. 498824.
6 Fiatal et al. Nicotine Tob Res 2016. 18(12): 2260-2267.
7 Nikolov et al. Drug Alcohol Depend 2011. 117(1): 62-5.
8 Borysovych et al. Biopolymers & Cell 2019. 35(5): 392.
9 Zahorodnia et al. Biopolymers & Cell 2019. 35(5): 393.
Vitalina Bashynska, PhD
Lab of Epigenetics, Institute of Gerontology NAMS Ukraine
Diagen genetic laboratory
Google Scholar
LinkedIn
ORCID
ResearchGate
Vitalina is the recipient of a small research grant from the US-Ukraine Foundation Biotech Initiative

