Thanks to a travel grant from the US-Ukraine Foundation, this summer I was lucky enough to participate in the Genome Engineering: CRISPR Frontiers conference, which is held annually at the Cold Spring Harbor Laboratory, USA.
However, this time, due to the coronavirus pandemic, the conference was held in a somewhat unusual format - online. But even in this format the conference was extremely interesting.
The research we presented at the poster session looked a bit modest compared to the other oral and poster presentations, but we’re new to this field and, more importantly, you can’t even compare our level of funding with that of teams from other countries working in the field of CRISPR.
All the oral presentations were delivered via Zoom. There were breakout rooms where participants could chat and discuss the sessions, and even an informal beer party.
Now about the content.
There were sessions on: CRISPR Biology, Technology, COVID, CRISPR Structure, DNA Repair, Plants and Therapeutics.
The conference started with a tribute to Rosalind Franklin. Fyodor Urnov delivered an emotional and interesting account of her life and work, and the significance of her famous photo #51 to understanding the structure of DNA.
Several other sessions were particularly interesting for me: COVID, Technology, CRISPR Structure Therapeutics.
CRISPR-based diagnostic systems were presented in the COVID session. These platforms aren’t limited to the SHERLOK test system, which has evolved into a one-step system, or the DETECTR or PAC-Man systems. In my opinion the development of these systems is a good example of how flexible the CRISPR tool is and how easily it adapts to different tasks. Among these systems, only the SHERLOK diagnostic kit was developed before the coronavirus pandemic; the others were developed quickly after COVID-19 appeared on earth.
My hero among the speakers in the Technology session was Audrone Lapinaite, who spoke about the development of editing systems, namely the version of the system that can convert adenine (A) into guanine (G). She spoke about the evolution of the TadA enzyme, which is responsible for these transitions in the structure of DNA. Their team finally found the answer to how TadA recognizes DNA as a substrate and why the ABE8 system (the latest generation) works so effectively. I also want to note that such base editors have colossal potential for the editing of pathogenic point mutations (or SNP) in clinical application. This approach is safer because it does not involve creating breaks in DNA.
It was also interesting to hear Martin Jinek talk about the structure of CRISPR, and specifically about the design and sequence of sgRNA, which is extremely important for developing a working tool with minimal off-target effects.
There were too many interesting lectures to list them all, but do I want to say that the progress with which this field is developing, including our knowledge of the diversity of CRISPR systems (almost every year new systems appear with interesting properties of Cas proteins) and the range of applications is impressive.
The conference organizers are leading authorities in the field of CRISPR:
Jennifer Doudna, University of California, Berkeley/HHMI
Maria Jasin, Memorial Sloan Kettering Cancer Center
David Liu, Broad Institute of Harvard and MIT
Jonathan Weissman, Whitehead Institute and MIT/HHMI
The conference will be held again next year, but whether it will be online or offline will depend on the situation in the world.
-Oksana Piven, Senior Scientist, Institute of Molecular Biology and Genetics NAS of Ukraine
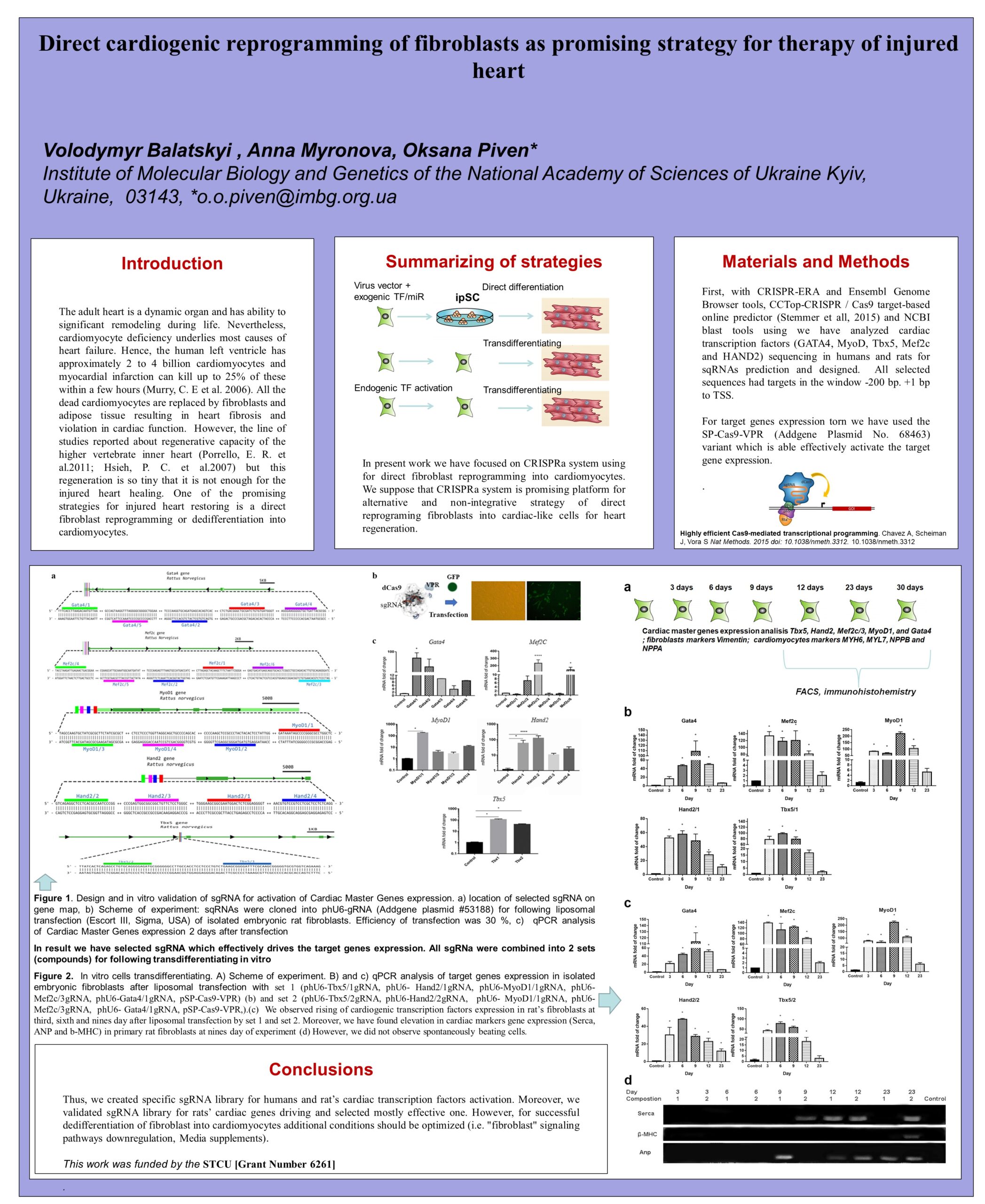
Read more about the research being done by Oksana and her colleagues to "roll back" a heart attack